Project Data
Genomic Answers for Kids
Advances in medicine happen more quickly when researchers share their findings and data. New studies build upon the published research of others, so it is essential that researchers publish their results in peer-reviewed scientific journals. Furthermore, generating new data is expensive and time consuming, so new findings are accelerated when researchers share data with each other. When many minds are looking at data, the chances for new insights increase. To support this, Genomic Answers for Kids is using new and innovative ways to share project data. Below you can find publications resulting from the project, ways that we share project data, and how researchers can request access to the data.
Publications
We publish our findings so they can be used to advance research and improve patient care.
- Spector BL, Yoo B, Miller N, Gaddis M, Thiffault I, Willig L. Association of rare variants in kidney developmental genes with chronic kidney disease and blood pressure: a UK Biobank study. Wis Med J. 123(1):e1-e8. Epub ahead of print. 2025 Mar 10.
- Chen X, Baker D, Dolzhenko E, Devaney J, Noya J, Berlyoung A, Brandon R, Hruska K, Lockovsky L, Kruszka P, Newman S, Farrow E, Thiffault I, Pastinen T, Kasperaviciute D, Gilissen C, Vissers L, Hoischen A, Berger S, Vilain E, Delot E, Eberle M, Genomics Research to Elucidate the Genetics of Rare Diseases Consortium (GREGoR). Genome-wide profiling of highly similar paralogous genes using HiFi sequencing. Nature Communications. 2025;16(1):2340.
- Groza C, Ge B, Cheung WA, Pastinen T*, Bourque G. Expanded methylome and quantitative trait loci detection by long-read profiling of personal DNA. Genome Research. In press. *Corresponding Author
- Rao A, Watt K, Mail L, Lamb M, Farrow E, Hassan H, Weaver K, Miller B, Dash S, Cox L, Gallacher L, Kant S, Gibson M, Pastinen T, Li D, Bhoj E, Zhu H, Zhang Y-B, Tan T, Trainor P, Cox T. Splicing defects and cell death cause SF3B2-linked craniofacial microsomia. Journal of Dental Research. In press.
- Means JC, Martinez-Bengochea AL, Louiselle DA, Nemechek JM, Perry JM, Farrow EG, Pastinen T, Younger ST. Rapid and scalable personalized ASO screening in patient-derived organoids. Nature. Published online January 22, 2025. DOI: 10.1038/s41586-024-08462-1.
- Chen X, Baker D, Dolzhenko E, Devaney JM, Noya j, Berlyoung AS, et al. Genome-wide profiling of highly similar paralogous genes using HiFi sequencing. Nature Communications. In press.
- Keskus A, Bryant A, Ahmad T, Yoo B, Aganezov S, et al. Severus: accurate detection and characterization of somatic structural variation in tumor genomes using long reads. Nature Biotechnology. In press.
- Gong M, Li J, Machado Bressan Wilke MV, Liu Y, Li Q et al. MARK2 variants cause autism spectrum disorder via the downregulation of WNT/β-catenin signaling pathway. Am J Hum Genet. 2024:S0002-9297(24)00366-5. Epub ahead of print. PMID: 39419027.
- Borroto MC, Patel H, Srivastava S, Swanson LC, Keren B, et al. Cohort Expansion and Genotype-Phenotype Analysis of RAB11A-Associated Neurodevelopmental Disorder. Pediatr Neurol. 2024;160:45-53. PMID: 39181022.
- Guitart X, Porubsky D, Yoo D, Dougherty ML, Dishuck P, et al. Independent expansion, selection and hypervariability of the TBC1D3 gene family in humans. Genome Res. 2024;gr.279299.124. Online ahead of print. PMID: 39107043
- Bakey Z, Cabrera OA, Hoefele J, Antony D, Wu K, et al. IFT74 variants cause skeletal ciliopathy and motile cilia defects in mice and humans. PLoS Genetics. 2023;19(6):e1010796. PMID: 37315079.
- Paul MS, Michener SL, Pan H, Chan H, Pfliger JM, et al. A syndromic neurodevelopmental disorder caused by rare variants in PPFIA3. Am J Hum Genet. 2024;111(1):96-118. PMID: 38181735
- Smail C, Ge B, Keever-Keigher MR, Schwendinger-Schreck C, Cheung WA, Johnston JJ, Barrett C, Genomic Answers for Kids Consortium, Feldman K, Cohen ASA, Farrow EG, Thiffault I, Grundberg E, Pastinen T. Complex trait Associations in rare diseases and impacts on Mendelian variant interpretation. Nature Communications.
- Cali E, Quirin T, Rocca C, Efthymiou S, Riva A, Marafi D, et al .Clinical and genetic delineation of autosomal recessive and dominant ACTL6B-related developmental brain disorders. Genetics in Medicine. 2024; doi: 10.1016/j.gim.2024.101251.
- Blackburn PR, Ebstein F, Hsieh T-C, Motta M, Radio FC, et al. Loss-of-Function Variants in CUL3 Cause a Syndromic Neurodevelopmental Disorder. Annals of Neurology. 2024; doi: 10.1002/ana/27077.
- Alstrup M, Cesca F, Krawczun-Rygmaczewska A, López-Menéndez C, Pose-Utrilla J, et al. Refining the phenotype of SINO syndrome: A comprehensive cohort report of 14 novel cases. 2024;26(11):101219.
- Perrier S, Macintosh J, Misiaszek AD, Lambert G, Guerrero K, et al. Novel Pathogenic Variants in POLR3K Cause POLR3-Related Leukodystrophy. Human Mutation. 2024;1:doi:10.1155/2024/8807171
- Furia F, Levy AM, Theunis M, Bamshad MJ, Bartos MN et al. The phenotypic and genotypic spectrum of individuals with mono- or biallelic ANK3 variants. Clinical Genetics. Online ahead of print.
- Rael VE, Yano JA, Huizar JP, Slayden LC, Weiss MA et al. Large-scale mutational analysis identifies UNC93B1 variants that drive TLR-mediated autoimmunity in mice and humans. Journal of Experimental Medicine. 2024;221( 8):e20232005. PMID: 38780621.
- Kalm T, Schob C, Völler H, Gardeitchik T, Gilissen C, Pfundt R, et al. Etiological involvement of KCND1 variants in an X-linked neurodevelopmental disorder with variable expressivity. American Journal of Human Genetics. 2024;111(6):1206-1221. PMID: 38772379.
- Mei Hong Liu, Benjamin Costa, Emilia C. Bianchini, Una Choi, Rachel C. Bandler, Emilie Lassen, Marta Grońska-Pęski, Adam Schwing, Zachary R. Murphy, Daniel Rosenkjær, Shany Picciotto, Vanessa Bianchi, Lucie Stengs, Melissa Edwards, Nuno Miguel Nunes, Caitlin A. Loh, Tina K. Truong, Randall E. Brand, Tomi Pastinen, J. Richard Wagner, Anne-Bine Skytte, Uri Tabori, Jonathan E. Shoag, Gilad D. Evrony. DNA mismatch and damage patterns revealed by single-molecule sequencing. Nature. In press
- Cohen ASA, Berrios CD, Zion TN, Barrett CM, Moore R, et al. Genomic Answers for Kids: Toward more equitable access to genomic testing for rare diseases in rural populations. American Journal of Human Genetics. 2024;111(5):825-832.
- Bhat S, Rousseau J, Michaud C, Lourenco CM, Stoler JM et al. Mono-allelic KCNB2 variants lead to a neurodevelopmental syndrome caused by altered channel inactivation. American Journal of Human Genetics. 2024;S0002-9297(24)00046-6
- De Pace R, Maroofian R, Paimboeuf A, Zamani M, Zaki MS, et al. Biallelic BORCS8 variants cause an infantile-onset neurodegenerative disorder with altered lysosome dynamics. Brain. 2024; 47(5):1751-1767.
- Groza C, Schwendinger-Schreck C, Cheung WA, Farrow EG, Thiffault I, Lake J, Rizzo WB, Evrony G, Curran T, Bourque G, and Pastinen T. Pangenome graphs improve the analysis of structural variants in rare genetic diseases. Nature Communications. 2023;15:657.
- Barrett C and Berrios C. Downstream Exclusion in Rural Rare Disease Precision Medicine Research. American Journal of Bioethics. In Press.
- Berrios C, McBeth M, Bradley-Ewing A, Schuetz N, Campbell A, et al. Developing a Community-Led Rare Disease ELSI Research Agenda. Orphanet Journal of Rare Diseases. 2024;19(1):23.
- Varberg KM, Dominguez EM, Koseva B, Varberg JM, McNally RP et al. Extravillous trophoblast cell lineage development is associated with active remodeling of the chromatin landscape. Nature Communications.2023;14(1):4826
- Farrow E, Jay A, Means J, Younger S, Biswell R et al. Case of CLPB deficiency solved by HiFi long read genome sequencing and RNA seq. American Journal of Medical Genetics: Part A. 2023;191(12):2908-2912.
- Liu Z, Xin B, Smith IN, Sency V, Szekely J et al. Hemizygous variants in protein phosphatase 1 regulatory subunit 3F (PPP1R3F) are associated with a neurodevelopmental disorder characterized by developmental delay, intellectual disability, and autistic features. Human Molecular Genetics. 2023;32(20):2981-2995.
- Bosch E, Popp B, Guse E, Skinner C, van der Sluijs PJ et al. Elucidating the clinical and molecular spectrum of SMARCC2-associated NDD in a cohort of 65 affected individuals. Genetics in Medicine. 2023;25(11):100950.
- Bhoj E, Ganapathi M, Matsuoka L, March M, Li D, et al. Heterozygous rare variants in NR2F2 cause a recognizable multiple congenital anomaly syndrome with developmental delays. European Journal of Human Genetics. 2023;31(10):1117-1124.
- Berrios C, Neal S, Zion T, Pastinen T. Comparing attitudes about genomic privacy and data sharing in adolescents and parents of children enrolled in a genomic research repository. American Journal of Bioethics: Empirical Bioethics. 2023;24:1-8.
- Calame DG, Guo T, Wang C, Garrett L, Jolly A et al. Monoallelic variation in DHX9, the gene encoding the DExH-box helicase DHX9, underlies neurodevelopment disorders and Charcot-Marie-Tooth disease. American Journal of Human Genetics. 2023;110(8):1394-1413. PMID:37467750
- Potic A, Perrier S, Radovic T, Gavrilovic S, Ostojic J, et al. Hypomyelination caused by a novel homozygous pathogenic variant in FOLR1: complete clinical and radiological recovery with oral folinic acid therapy and review of the literature. Orphanet Journal of Rare Diseases. 2023;18(1):187. PMID:37443037
- Strenk ME, Berrios C, Garrett JR. Addressing the burdens that newborn screening imposes on underserved communities. American Journal of Bioethics. 2023;23(7):79-82. PMID: 37339296.
- McCormick EM, Keller K, Taylor JP, Coffey AJ, Shen L, et al. Expert panel curation of 113 primary mitochondrial disease genes for the Leigh syndrome spectrum. Ann Neurol. 2023;94(4):696-712. PMID: 37255483.
- Macintosh J, Thiffault I, Pastinen T, Sztriha L, and Bernard G. A recurrent de novo variant in EIF2AK2 causes a hypomyelinating leukodystrophy. Child Neurol Open. 2023;eCollection: 10:2329048X231176673. PMID: 37284702
- Perrier S, Guerrero K, Tran LT, Michell-Robinson MMA, Legault G, et al. Solving inherited white matter disorder etiologies in the neurology clinic: Challenges and lessons learned using next-generation sequencing. Frontiers in Neurology. 2023; 14:1148377.
- Calame DG, Moreno VC, Berger S, Lotze T, Shinawi M, et al. Cation leak through the ATP1A3 pump causes spasticity and intellectual disability. Brain. 2023;146(8):3162-3171. PMID:37043503.
- Mirchi A, Guay SP, Tran LT, Wolf NI, Vanderver A, Brais B, et al. Craniofacial features of POLR3-related leukodystrophy caused by allelic variants in POLR3A, POLR3B and POLR1C. Journal of Medical Genetics. 2023;60(10):1026-1034.
- Kane NJ, Cohen ASA, Berrios C, Jones B, Pastinen T, Hoffman MA. (2023) Committing to Genomic Answers for All Kids: Evaluating Inequity in Genomic Research Enrollment. Genetics in Medicine. 2023;25(9):100895. PMID: 37194653.
- Tucker M, Yu W, Menden H, Xia S, Schwendinger-Schreck C, et al. IRF7 and UNC93B1 variants in an infant with recurrent herpes simplex virus infections. Journal of Clinical Investigation. 2023;133(11):e154016. PMID:37097753.
- Cheung WA, Johnson AF, Rowell WJ, Farrow E, Hall R, et al. Direct haplotype-resolved 5-base HiFi sequencing for genome-wide profiling of hypermethylation outliers in a rare disease cohort. Nature Communications. 2023;14(1):3090. PMID: 37248219.
- Chen X, Harting J, Farrow E, Thiffault I, Kasperaviciute D et al. Comprehensive SMN1 and SMN2 profiling for spinal muscular atrophy analysis using long-read PacBio HiFi sequencing. American Journal of Human Genetics. 2023;110(2):240-250.
- Hiatt SM, Trajkova S, Sebastiano MR, Partridge EC, Abidi FE, et al. Deleterious, protein-altering variants in the X-linked transcriptional coregulator ZMYM3 in 22 individuals with a neurodevelopmental delay phenotype. American Journal of Human Genetics. 2023;110(2):215-227.
- Zion TN, Berrios CD, Cohen ASA, Bartik L, Cross LA et al. Insurance denails and diagnostic rates in a pediatric genomic research cohort. Genetics in Medicine. 2023:25(5):100020.
- do Rosario MC, Bey GR, Nmezi B, et al. Variants in the zinc transporter TMEM163 cause a hypomyelinating leukodystrophy. Brain. 2022:145(12): 4202-4209. PMID: 35953447
- McQuerry JA, Mclaird M, Hartin SN, Means JC, Johnston C, Pastinen T, Younger ST. Massively parallel identification of functionally consequential noncoding genetic variants in undiagnosed rare disease patients. Sci Rep. 2022. May 9;12(1):7576. PMID: 35534523
- Berrios C, Sadaro S, Sandritter T, et al. Parental understanding and attitudes following pharmacogenomic testing for pediatric neuropsychiatric patients. Pharmacogenomics. 2022;23(6):345-354. PMID: 35311353
- Goldman JL, Miller JO, Miller N, et al. HLA-B*07:02 and HLA-C*07:02 are associated with trimethoprim-sulfamethoxazole respiratory failure. Pharmacogenomics J. 2022;22(2):124-129. PMID: 35169303
- Cohen ASA, Farrow EG, Abdelmoity AT, Alaimo JT, Amudhavalli SM, et al. Genomic answers for children: dynamic analyses of >1000 pediatric rare disease genomes. Genet Med. 2022;24(6):1336-1348. PMID: 35305867
- Helman G, Mendes MI, Nicita F, Darbelli L, Sherbini O, et al. Expanded phenotype of AARS1-related white matter disease. Genet Med. 2021;23(12):2352-2359. PMID: 34446925
- Bruel AL, Vitobello A, Thiffault I, Manwaring L, Willing M, et al. ITSN1: a novel candidate gene involved in autosomal dominant neurodevelopmental disorder spectrum. Eur J Hum Genet. 2022;30(1):111-116. PMID: 34707297
- Von der Lippe C, Tveten K, Prescott TE, Holla OL, Busk OL, et al. Heterozygous variants in ZBTB7A cause a neurodevelopmental disorder associated with symptomatic overgrowth of pharyngeal lymphoid tissue, macrocephaly, and elevated fetal hemoglobin. Am J Med Genet. 2022;188(1):272-282. PMID: 34515416
- Berrios C, Hurley EA, Willig L, Thiffault I, Saunders C, Pastinen T, Goggin K, Farrow E. (2021) Challenges in genetic testing: clinician variant interpretation processes and the impact on clinical care. Genet Med. 2021:23(12):2289-2299. PMID: 34257423
- Gillentine MA, Wang T, Hoekzema K, Rosenfeld J, Peng-fei L, et al. Rare deleterious mutations of HNRNP genes result in shared neurodevelopmental disorders. Genome Medicine. 2021;13(1):63. PMID: 33874999
- Kunova-Bosakova M, Abraham SP, Nita A, Hruba E, Buchtova M et. al. Mutations in GRK2 cause Jeune syndrome by impairing Hedgehog and canonical Wnt signaling. EMBO Molecular Medicine. 2020;12(11):e11739. PMID: 33200460
- Hartin S, Means JC, Alaimo JT, and Younger ST. Expediting rare disease diagnosis: a call to bridge the gap between clinical and functional genomics. Mol Med. 2020;26(1):117. PMID: 33238891
- Jenkins J, Barnes A, Birnbaum B, Papagiannis J, Thiffault I, et. al. LZTR1-Related Hypertrophic Cardiomyopathy Without Typical Noonan Syndrome Features. Circ Genom Precis Med. 2020;13(2):e002690. PMID: 32004086
- den Hoed J, de Boer E, Voisin N, Dingemans AJM, Guex N, et al. Mutation-specific pathophysiological mechanisms define different neurodevelopmental disorders associated with SATB1 dysfunction. Am J Hum Gen. 2021;108(2):346-356. PMID: 33513338
- Connaughton DM, Dai R, Owen DJ, Marquez J, Mann N, et al. Mutations of the transcriptional corepressor ZMYM2 cause syndromic urinary tract malformations. Am J Hum Genet. 2020; 107(4):727-742. PMID: 32891193
- Kummeling J, Stremmelaar DE, Raun N, Reijnders MRF, Willemsen MH, et al. Characterization of SETD1A haploinsufficiency in humans and Drosophila defines a novel neurodevelopmental syndrome. Mol Psychiatry. 2021; 26(6):2013-2024. PMID: 32346159
- Mao D, Reuter CM, Ruzhnikov MRZ, Beck AE, Farrow EG, et al. De novo EIF2AK1 and EIF2AK2 Variants Are Associated with Developmental Delay, Leukoencephalopathy, and Neurologic Decompensation. Am J Hum Genet. 2020;106(4):570-583. PMID: 32197074
- Chilton I, Okur V, Vitiello G, Selicorni A, Mariani M, et al. De novo heterozygous missense and loss-of-function variants in CDC42BPB are associated with a neurodevelopmental phenotype. Am J Med Genet A. 2020;182(5):962-973. PMID: 32031333
- Yan K, Rousseau J, Machol K, Cross LA, Agre KE, et al. Deficient histone H3 propionylation by BRPF1-KAT6 complexes in neurodevelopmental disorders and cancer. Sci Adv. 2020; 6(4):eaax0021. PMID: 32010779
- Scott TM, Guo H, Eichler EE, Rosenfeld JA, Pang K, et al. BAZ2B haploinsufficiency as a cause of developmental delay, intellectual disability, and autism spectrum disorder. Hum Mutat. 2020:41(5);921-925. PMID: 31999386
- Li L, Ghorbani M, Weisz-Hubshman M, Rousseau J, Thiffault I, et al. Lysine acetyltransferase 8 is involved in cerebral development and syndromic intellectual disability. J Clin Invest. 2020; 130(3):1431-1445. PMID: 31794431
- Donkervoort S, Sabouny R, Yun P, Gauquelin L, Chao KR, et al. MSTO1 mutations cause mtDNA depletion, manifesting as muscular dystrophy with cerebellar involvement. Acta Neuropathol. 2019;138(6):1013-1031. PMID: 31463572
- Patak J, Gilfert J, Byler M, Neerukonda V, Thiffault I, et al. MAGEL2-related disorders: A study and case series. Clin Genet. 2019;96(6):493-505. PMID: 31397880
- Jeong H, Dishuck PC, Yoo D, Harvey WT, Munson KM, Lewis AP, Kordosky J, Garcia GH; Human Genome Structural Variation Consortium (HGSVC); Yilmaz F, Hallast P, Lee C, Pastinen T, Eichler EE. Structural polymorphism and diversity of human segmental duplications. bioRxiv [Preprint]. 2024 Jun 6:2024.06.04.597452. doi: 10.1101/2024.06.04.597452.PMID: 38895457
- Groza C, Ge B, Cheung W, Pastinen T, Bourque G. Expanded methylome and quantitative trait loci detection by long-read profiling of personal DNA. bioRxiv
Data Sharing
We share our data with the scientific community through several mechanisms designed to advance research in pediatric disease, while protecting participants’ confidentiality. Summary data on variants is publicly available, while individual level data is under controlled access and available only to other researchers studying pediatric genetic disease.
Summary Level Data
Variants identified in Genomic Answers for Kids may be searched using the Beacon Network. The Beacon Network is an initiative of the Global Alliance for Genomics and Health that aggregates data for worldwide genomic data sets and allows searches for variants across data sets. The Genomic Medicine Center also shares variant level data with ClinVar a National Library of Medicine hosted database of genomic variants that includes information about disease associations and laboratory interpretations of variant pathogenicity.
PhenoTips®
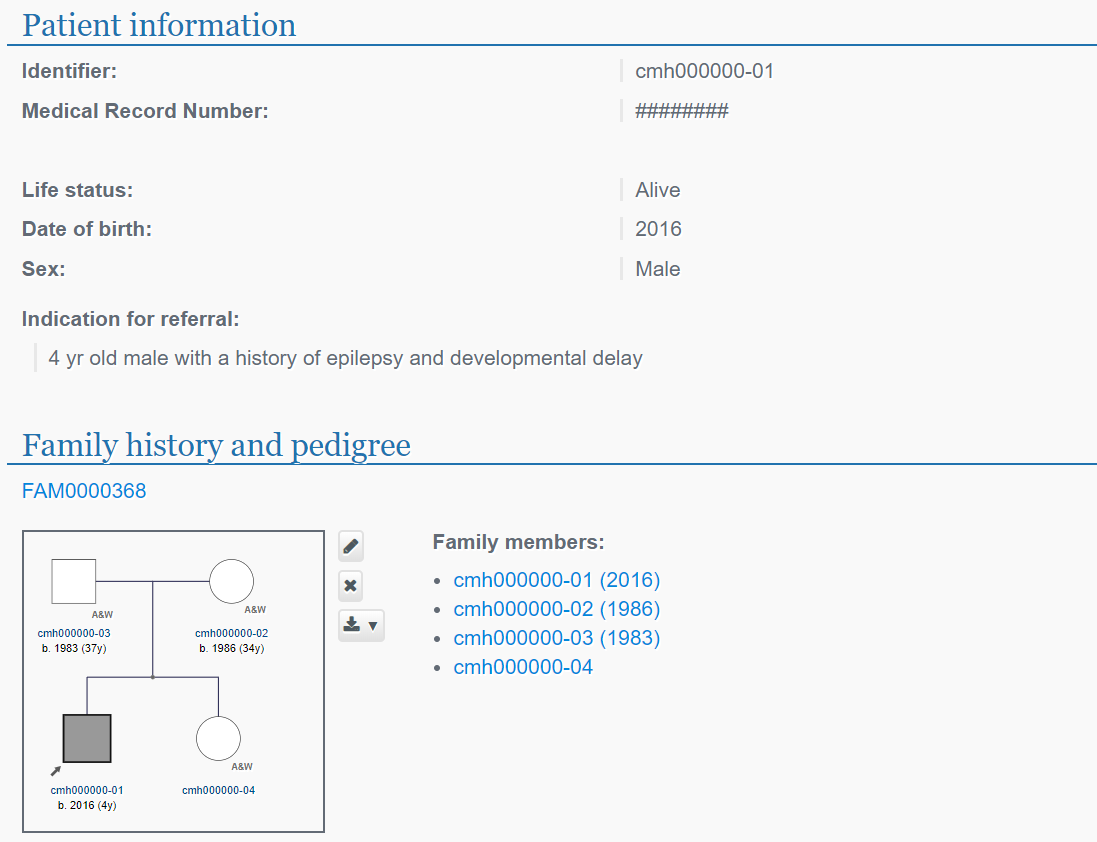
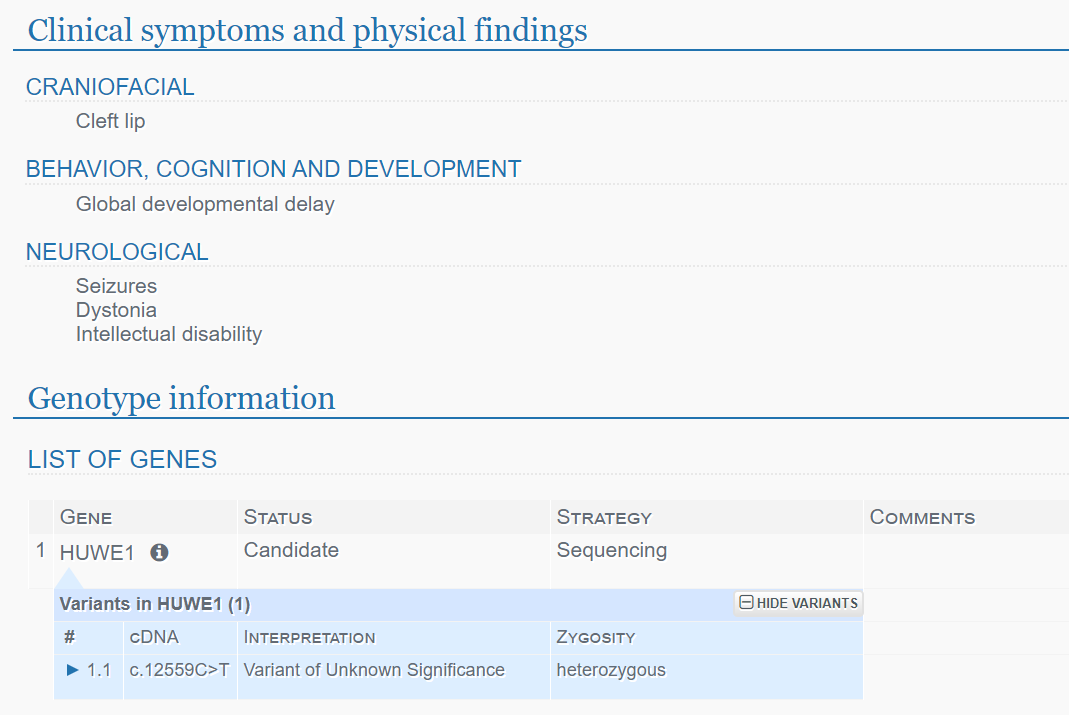
Individual level data including prioritized variants, patient symptoms and phenotype, putative diagnoses and diagnostic variants are available through a controlled access version of the PhenoTips® software. Individuals’ symptoms are entered using the Human Phenotype Ontology (HPO) with diagnoses stored using Online Mendelian Inheritance in Man (OMIM) phenotype identifiers. Individuals’ symptoms can be exported in a simple delimited text format for analysis and can be integrated in a number of open source tools and algorithms for automated diagnosis and candidate gene nomination. Prioritized, annotated variant lists for each individual are provided with display options to show genomic annotation and cross-referencing to external databases. The database can be searched by phenotype, diagnosis and candidate gene with case level details presented for each individual.
Database of Genotypes and Phenotypes (dbGaP)
Individual level genomic data including raw sequencing data, aligned reads, SNVs, small insertion/deletions and structural variant calls will be periodically deposited, along with phenotype data, into the NIH Database of Genotypes and Phenotypes (dbGaP). Access to the data is controlled by dbGaP and follows the conditions of the study’s informed consent. The data in dbGaP will also be available for analysis using the NHGRI Genomic Data Science Analysis Visualization and Informatics Lab-space (AnVIL).
Information on the Genomic Answers for Kids data available in dbGaP are available at the National Center for Biotechnology Information website.
Learn more and request access
Pediatric researchers may request access to individual level data from the Genomic Answers for Kids program.